Jwala Dhamala
I am a Senior Scientist at Amazon AGI, California. My research focuses on advancing Artificial Intelligence through the development of large language models, agentic models, and reasoning models that are helpful, capable, and safe. My specific interests include benchmark curation, the design of robust evaluation metrics, and the evaluation of models to assess their alignment with responsible AI policies. I am also engaged in uncovering model vulnerabilities through novel jailbreak attacks and red-teaming methodologies.
Prior to joining Amazon, I completed my Ph.D. in Computing and Information Sciences at the Rochester Institute of Technology (RIT), where I worked under the supervision of Dr. Linwei Wang in the Computational Biomedicine Lab. My doctoral research centered on personalization and uncertainty quantification in multi-scale 3D simulation models of cardiac electrophysiology. This work allowed me to operate at the intersection of machine learning—specifically Bayesian modeling, optimization, generative modeling, and graph convolutional networks—and computational healthcare, with a focus on personalized cardiac modeling.
News
Earlier News (2020 and before)
- [Dec, 2020] Panelist on AI fairness discussion at NeurIPS 2020: Watch here.
- [Oct, 2020] Paper with intern Ansel accepted at EMNLP workshop.
- [Feb, 2020] Paper accepted at Medical Image Analysis.
- [Feb, 2020] Successfully defended PhD: Thesis.
- [Dec, 2019] Joined Amazon Alexa NU-AI as Research Scientist.
- [Jun, 2019] Paper finalist for MICCAI Young Scientist Award.
- [Oct, 2018] Paper accepted at IEEE Sensors Letters and NeurIPS ML4H.
- [Oct, 2018] Abstract accepted to WiML Workshop 2018.
- [Sep, 2018] Paper finalist for MICCAI 2018 Young Scientist Award.
Selected Publications
For a comprehensive list of my publications, please visit my Google Scholar profile.Conference Publications
-
MICo: Preventative Detoxification of Large Language Models through Inhibition ControlNAACL Findings, 2024
-
Tokenization Matters: Navigating Data-Scarce Tokenization for Gender Inclusive Language TechnologiesNAACL Findings, 2024
-
Tree-of-Traversals: A Zero-Shot Reasoning Algorithm for Augmenting Black-box Language Models with Knowledge GraphsACL, 2024
-
“I’m fully who I am”: Towards Centering Transgender and Non-Binary Voices to Measure Biases in Open Language GenerationFAccT, 2023
-
Resolving Ambiguities in Text-to-Image Generative ModelsACL, 2023
-
Mitigating Gender Bias in Distilled Language Models via Counterfactual Role ReversalACL Findings, 2022
-
On the Intrinsic and Extrinsic Fairness Evaluation Metrics for Contextualized Language RepresentationsACL, 2022
-
BOLD: Dataset and Metrics for Measuring Biases in Open-Ended Language GenerationACM FAccT, 2021
-
Bayesian Optimization on Large Graphs via a Graph Convolutional Generative Model: Application in Cardiac Model PersonalizationMICCAI, 2019
-
High-dimensional Bayesian Optimization of Personalized Cardiac Model Parameters via an Embedded Generative ModelMICCAI, 2018
-
Quantifying the Uncertainty in Model Parameters using Gaussian Process-Based Markov Chain Monte Carlo: An Application to Cardiac Electrophysiological ModelsIPMI, 2017
-
Spatially-Adaptive Multi-scale Optimization for Local Parameter Estimation: Application in Cardiac Electrophysiological ModelsMICCAI, 2016
Journal Publications
-
Embedding High-dimensional Bayesian Optimization via Generative Modeling: Parameter Personalization of Cardiac Electrophysiological ModelsMedical Image Analysis (MedIA), in submission
-
Multivariate Time-series Similarity Assessment via Unsupervised Representation Learning and Stratified Locality Sensitive Hashing: Application to Early Acute Hypotensive Episode DetectionIEEE Sensors Letters, 2018; NeurIPS ML4H Workshop, 2018
-
Quantifying the Uncertainty in Model Parameters using Gaussian Process-Based Markov Chain Monte Carlo in Cardiac ElectrophysiologyMedical Image Analysis (MedIA), 2018
-
Spatially-Adaptive Multi-Scale Optimization for Local Parameter Estimation in Cardiac ElectrophysiologyIEEE Transactions on Medical Imaging (TMI), 2017
Selected Projects
We introduce BOLD, a large-scale dataset of 23,679 English prompts to benchmark social biases in open-ended text generation across five domains: profession, gender, race, religion, and political ideology. We also propose automated metrics for toxicity, psycholinguistic norms, and gender polarity. Analysis of outputs from three popular language models shows they exhibit greater bias than human-written Wikipedia text across all domains.
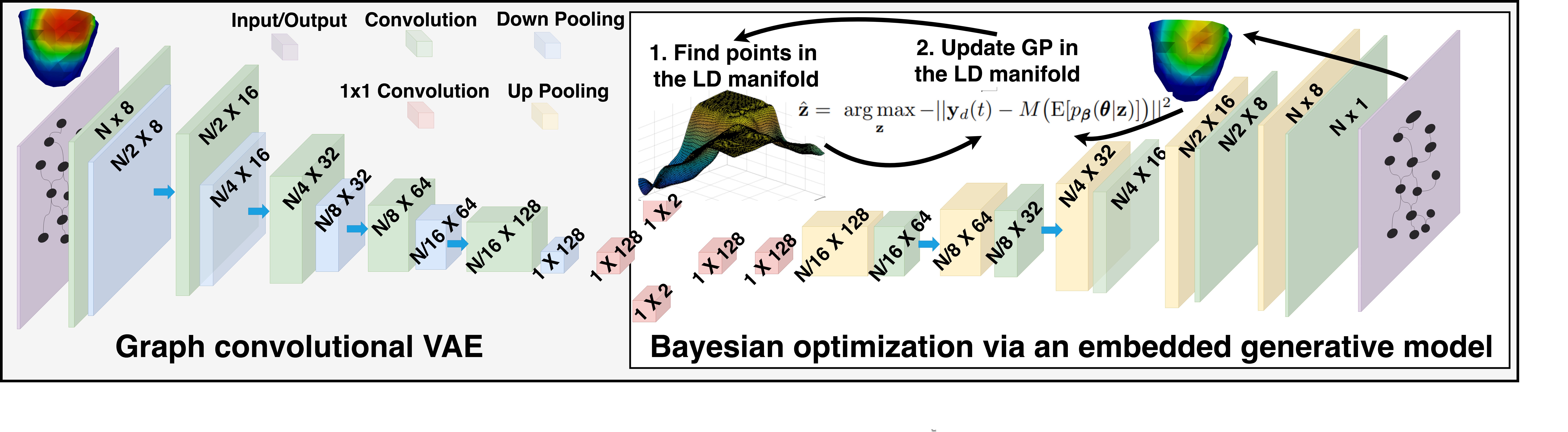
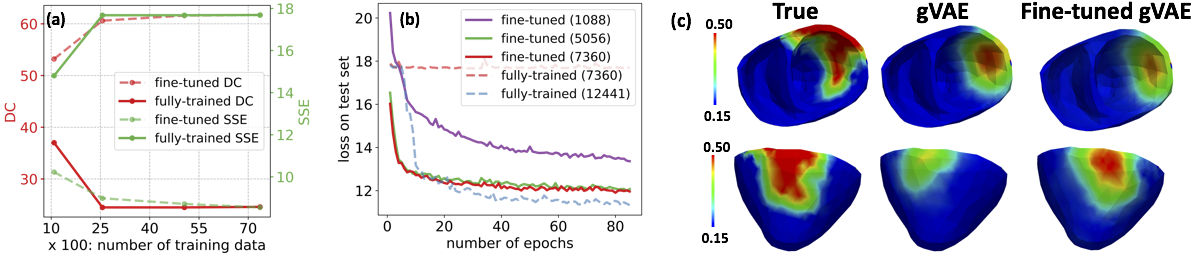
We present a novel graph convolutional VAE to allow generative modeling of non-Euclidean data, and utilize it to embed Bayesian optimization of large graphs into a small latent space. This approach bridges the gap of previous works by introducing an expressive generative model that is able to incorporate the knowledge of spatial proximity and hierarchical compositionality of the underlying geometry. It further allows transferring of the learned features across different geometries.
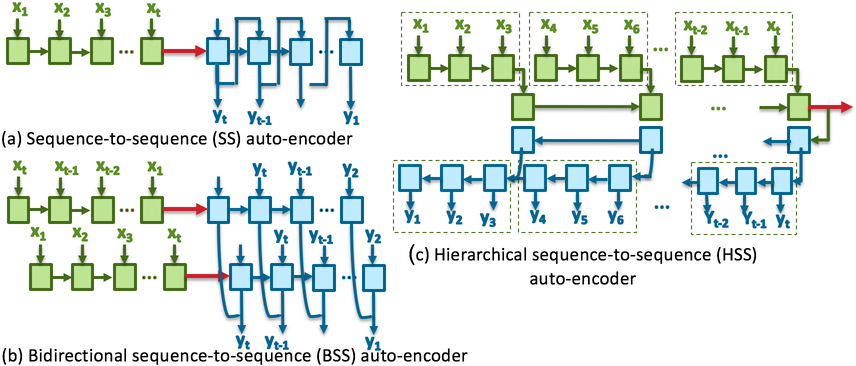
We learn the representations of multivariate time-series of physiologic signals with a sequence-to-sequence auto-encoder. We then hash the learned multivariate time-series representations of labeled dataset to enable signal similarity assessment. This methodological framework is evaluated to predict Acute Hypotensive Episodes (AHE) on vital signal recordings extracted from eICU Collaborative Research Database.
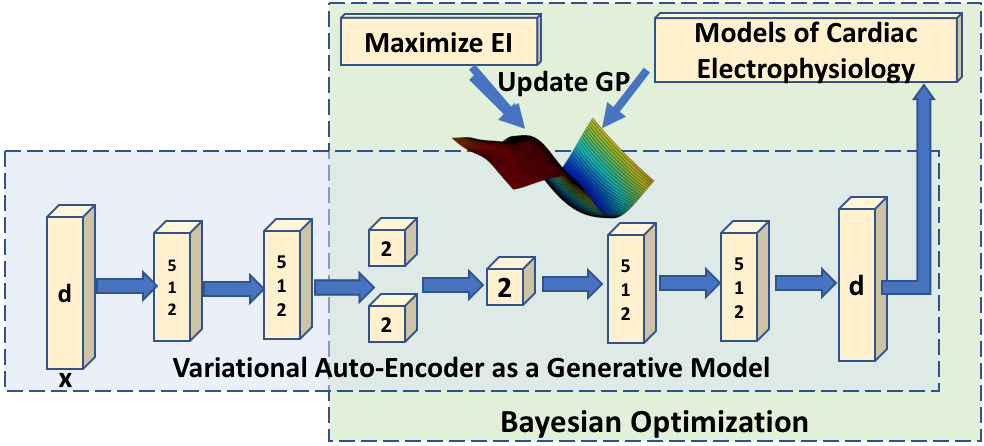
We devise a novel concept that embeds a generative variational auto-encoder (VAE) into the objective function of Bayesian optimization, providing an implicit low-dimensional (LD) search space that represents the generative code of the HD spatially-varying tissue properties. In addition, the VAE-encoded knowledge about the generative code is used to guide the exploration of the search space. It is applied to estimating high-dimensional tissue excitability in a cardiac electrophysiological model.
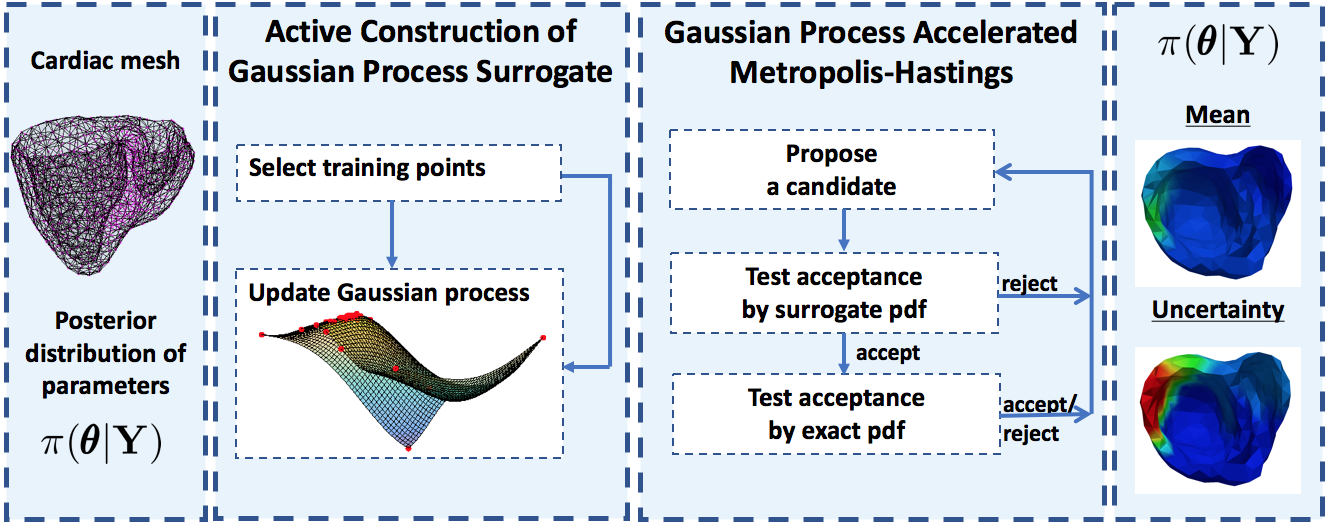
The quantification of uncertainty in model parameters is challenging because the posterior distribution of the parameters given the measurement data is non-Gaussian and the evaluation of the model is computationally expensive. In this project, we l earn a surrogate of this complicated and computationally expensive posterior distribution and utilize it to obtain a MCMC sampling with higher acceptance rate. The surrogate posterior pdf is used to accelerate the sampling of the true posterior pdf and not as a replacement.
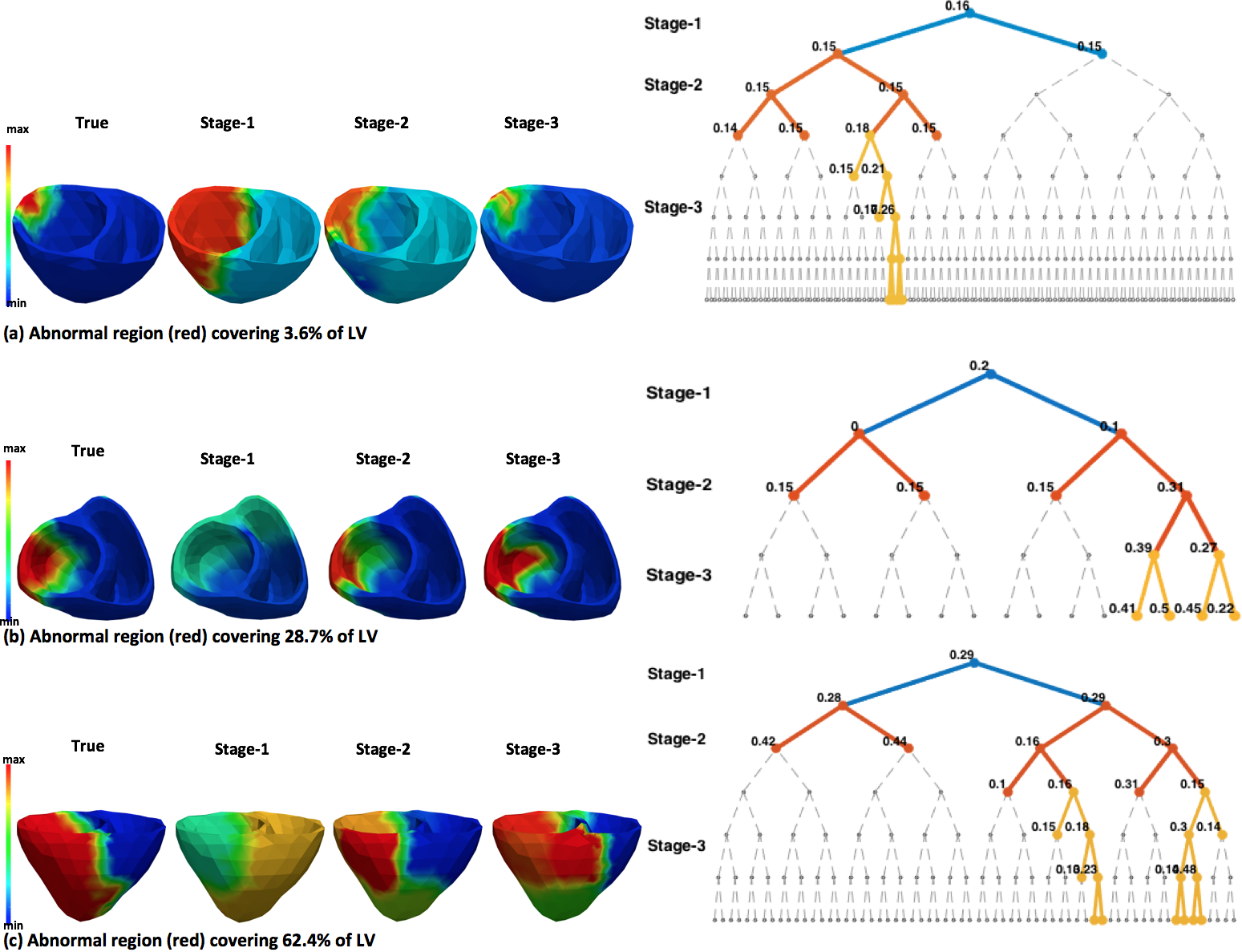
We present a novel framework that, going beyond a uniform low-resolution approach, is able to obtain a higher resolution estimation of tissue properties represented by spatially non-uniform resolution. This is achieved by two central elements: 1) a multi-scale coarse-to-fine optimization that facilitates higher resolution optimization using the lower resolution solution, and 2) a spatially adaptive decision criterion that retains lower resolution in homogeneous tissue regions and allows higher resolution in heterogeneous tissue regions. The presented framework is evaluated in estimating the local tissue excitability properties of a cardiac EP model.
Activities
Workshop link: trustnlpworkshop.github.io
Workshop link: Responsible AI at KDD 2021
Relevant Publications:
[1] Sandesh Ghimire, Jwala Dhamala, et al. “Overcoming Barriers to Quantification and Comparison of Electrocardiographic Imaging Methods...” Computing in Cardiology, IEEE, 2017 (To appear).
[2] Jaume Coll-Font*, Jwala Dhamala, et al. “The Consortium on Electrocardiographic Imaging.” CINC, 2016.